Google matrix analysis
of bi-functional SIGNOR network
of protein-protein interactions
by K.M.Frahm and D.L.Shepelyansky
bioRxiv:https://doi.org/10.1101/750695


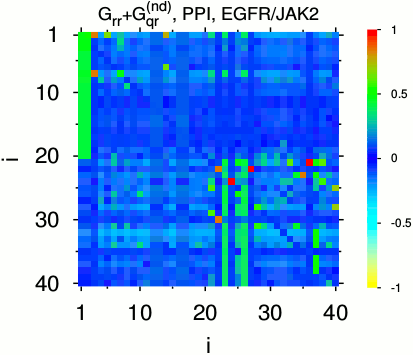
Left: matrix elements (color) of
two components of reduced Google matrix
of SIGNOR PPI network obtained for 40 most sensitive nodes on the pathway
from protein EGFR P00533 (+) to protein JAK2 O60674 (-).
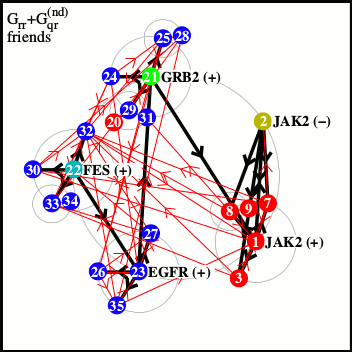
Right: network of friends of most significant nodes from left panel.
| |
 Article download:
bioRxiv-v1 (here) Article download:
bioRxiv-v1 (here) All figs in gzip format
(here) All figs in gzip format
(here)  All network figs
for components of reduced Google matix of 40 LIRGOMAX proteins: All network figs
for components of reduced Google matix of 40 LIRGOMAX proteins:injection at EGFR P00533 (+) and absorption at JAK2 O60674 (-) of Table-1 (here); response amplitudes (P_1) with names and ranks of proteins (here) gziped directory of files of matrix elements (here), readme file for gziped directory (here )  All network figs
for components of reduced Google matix of 40 LIRGOMAX proteins All network figs
for components of reduced Google matix of 40 LIRGOMAX proteinsfor injection at MAP2K1 Q02750 (+) and absorption at EGFR P00533 (-) of Table-3 (here); response amplitudes (P_1) with names and ranks of proteins (here) gziped directory of files of matrix elements (here), readme file for gziped directory (here)  All network figs
for components of reduced Google matix of 40 LIRGOMAX proteins All network figs
for components of reduced Google matix of 40 LIRGOMAX proteinsfor injection at MAP2K1 Q02750 (+) and absorption at EGFR P00533 (-) of Table-4 (here); response amplitudes (P_1) with names and ranks of proteins (here) gziped directory of files of matrix elements for Table-4 (here), gziped directory of files of matrix elements with $i=20$ being 20th most negative protein value of P_1 (see paper) (here), readme file for gziped directory (here)  PageRank of proteins
(single functional links
(here))
and (bi-functional links (here)); PageRank of proteins
(single functional links
(here))
and (bi-functional links (here));gziped directory of PageRank, CheiRank, magnetization and README files is (here )  PageRank magnetization of all proteins
is (here) PageRank magnetization of all proteins
is (here) Executive code
to obtain LIRGOMAX and REGOMAX results (as examples above) on Executive code
to obtain LIRGOMAX and REGOMAX results (as examples above) onPC-computers under Linux (see README files) with an example of results (here)  LIRGOMAX algorithm
is explained in detail at
(web site); LIRGOMAX algorithm
is explained in detail at
(web site);REGOMAX algorithm is explained in detail at (web site)
This webpage is created at August 27, 2019 and is maintained
|